Genomic Basis of Fish Biodiversity
With more than 30,000 living species, ray-finned fishes (Actinopterygii) are the largest group of vertebrates. Their biodiversity is concentrated in the teleost fishes that include >96% of extant actinopterygians. Our research aims to uncover the genomic secrets of the teleost evolutionary success.
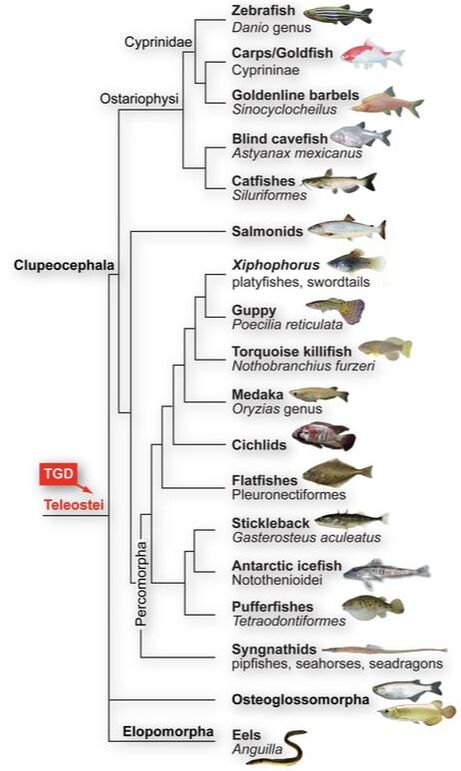
Teleost fishes are derived from a lineage-specific whole genome duplication (TGD) and we are studying the connection between this 'genomic big bang' and the genomic and phenotypic diversification of teleosts. We are involved in the comparative genomic analyses of a broad variety of fish genomes (e.g. coealcanth, platyfish, sturgeon). In collaboration with the Broad Institute, we have sequenced and analyzed the genome of the spotted gar (Lepisosteus oculatus). Gar is an emerging model system for comparative genomics and serves as ‘unduplicated’ ray-finned fish outgroup to teleosts as well as to lobe-finned fishes including tetrapods. The gar genome is a good representative of the ancestral bony vertebrate genome and has retained many old genes that have been lost in more derived lineages such as teleosts and tetrapods. Evolution by Gen[om]e DuplicationWe aim to develop a comprehensive understanding of the evolutionary consequences of whole genome duplications (WGD). WGDs occurred early in vertebrate evolution and are particularly common among fishes, e.g. in the modern teleost fishes such as zebrafish and medaka. Importantly, WGDs are thought to provide extra genetic material for major evolutionary transitions, for the emergence of novel morphologies, and to facilitate biodiversification.
Understanding the evolutionary consequences of these WGDs is critical to (1) evaluate the contribution of WGDs to vertebrate key innovations and diversification (2) understand disease genes in our own genome since many human genetic disorders involve WGD-derived genes (2) improve our understanding of the relationships among vertebrate genomes that are crucial for translating biomedical results from zebrafish to humans |
Evolution of Gene RegulationWe investigate gene regulation in fishes in comparison to other vertebrates, including humans. Such analyses are challenged by extensive sequence divergence between teleost fishes and other vertebrates which particularly hinders sequence comparisons in regions of the genome that regulate gene functions. However, such comparisons are essential to translate genetic knowledge from zebrafish to human and to evaluate modes of gene expression evolution following gene duplication. At the genome sequence level, gar and bowfin can serve as links among divergent vertebrate genomes by ‘bridging’ teleost (e.g. zebrafish) to human gene regulatory elements. We are using comparative genomics to identify conserved non-coding elements (CNEs, often functional sites in the genome) and use novel epigenomic profiling techniques (ATAC-seq, HiChIP) to identify in vivo the gene regulatory landscape during the course of fish embryonic development and in adult tissues.
This work is supported by a grant from NIH ORIP to establishing genome-wide catalogues of regulatory elements in fishes to connect their gene regulation with human disease data. |
Advancing Comparative Medicine and Evo-Devo with New Fish Models
Zebrafish and other teleost fishes are favorable research models that can be leveraged for biomedical purposes. However, establishing genome relationships from teleosts to humans is difficult because they represent highly derived vertebrate lineages. Using non-teleost fishes such as gar and bowfin as ‘bridge species’, we can connect biomedical information gathered from teleosts to human biology and disease. Our group has established a probably world-wide unique Gar Facility at MSU.
Gar is an excellent model for studying the evolution of vertebrate development due to its archaic body plan: for example, our studies in gar illuminate the evolution of pigment patterns and of the skeletal system. Fin regeneration is more pronounced in gar than in zebrafish and we establish gar as a new model to study the genetics of regeneration. We are developing functional genetic techniques in gar and - supported by an NSF EDGE grant - establish CRISPgaR genome engineering and transgenesis. |
|
|
Evolution and Development of
|