Welcome to the Fish Evo Devo Geno Lab!
|
Our research focuses on genomic and morphological change in vertebrates over long macro-evolutionary timescales. We utilize diverse groups of fishes such as gar, bowfin, zebrafish, medaka, and killifishes as model systems to answer fundamental questions about the developmental and genomic basis of major transitions during vertebrate animal evolution, including our own human lineage.
Our integrative research program combines comparative genomics with experimental developmental studies of gene functions to answer questions that have long interested evolutionary biologists: (1) How does biodiversity arise from diversification of genome content by gen(om)e duplication and gene loss? (2) How do changes in gene regulation contribute to evolutionary novelties and key innovations? (3) How can evolutionary genetic principles be applied to improve the understanding of human disease? |
Lab News
2023
October 2023
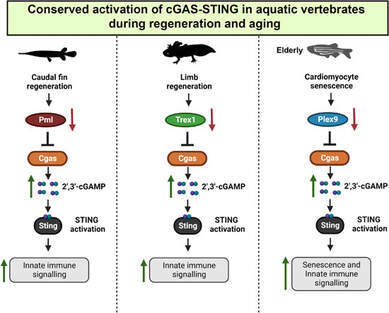
New publication out in Journal of Experimental Zoology B! This collaborative study with Saby Mathavarajah, Graham Dellaire, and colleagues shows that the cellular cGAS-STING response is conserved across gar fin and axolotl limb regeneration and zebrafish ageing. This work is part of the upcoming JEZB Special Issue "Aquatic Models for Biomedical Evo-Devo".
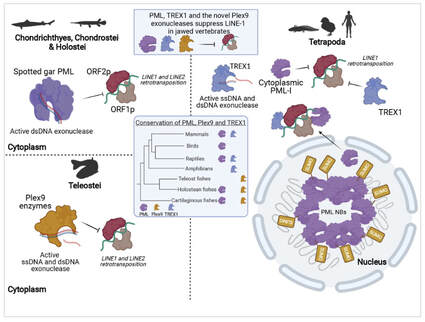
The study complements a paper with Saby and Graham on genome maintenance and retroelement control including our spotted gar (and a gar cell line!) published earlier this year in Nucleic Acids Research.
The study complements a paper with Saby and Graham on genome maintenance and retroelement control including our spotted gar (and a gar cell line!) published earlier this year in Nucleic Acids Research.
October 2023
|
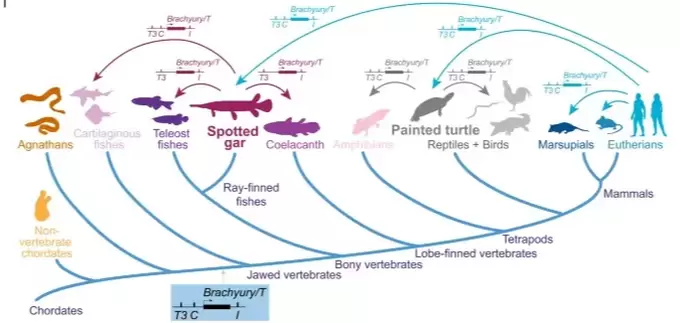
The "Gar Bridge" does it again! In this international collaboration led by the Mosimann-Burger and Kozmik labs published in Nature Communications, we show that the slowly evolving gar genome identifies hidden sequence and functional conservation of enhancer elements from tetrapods to fishes around the iconic Brachyury/T/Tbxtb transcription factor gene. Brachyury is essential to make the notochord, the embryonic stiffening structure that gives our phylum Chordata its name... now we know how!
|
July 2023
|
Our gars make it into PNAS! Read this comparative and highly collaborative study by Jan Stundl et al. to learn how the neural crest gave rise to ancient vertebrate dermal armor and see the scales of gar and other fishes spin.
See also the commentary by Andrew Gillis in PNAS. |
|
May 2023
Award-winning undergraduate researchers Keyana Blake and Daniel Do graduate from MSU - congratulations!
Keyana will join the IPREP postbac program and Daniel follows former postdoc, now faculty, Drew Thompson to join Drew's Xtremo-Devo Lab at Western Michigan as Master's student.
Keyana will join the IPREP postbac program and Daniel follows former postdoc, now faculty, Drew Thompson to join Drew's Xtremo-Devo Lab at Western Michigan as Master's student.
Spring 2023
We have recruited several new undergraduate researchers: Alexandra Stapleton, Isabella Rinaldi, and Grace Urban.
February 2023
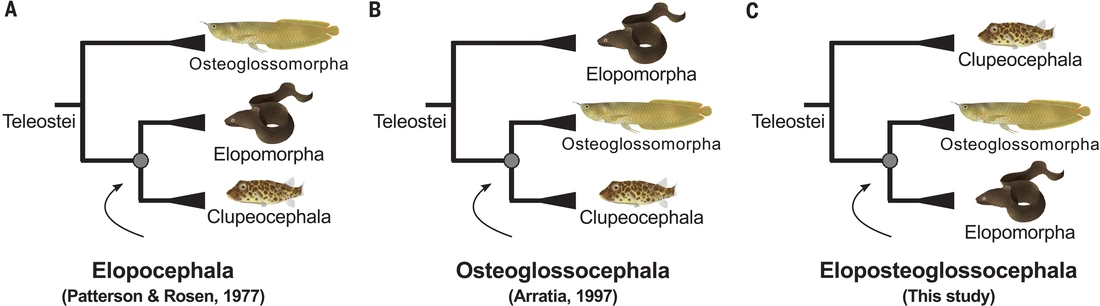
We are glad to have contributed to the GenoFish Consortium that used genome structure analyses of many new fish genomes to resolve one of the most stubborn nodes of teleost fish phylogeny. After decades long debates, it turns out that the previously least suspicious solution is the one: Elopomorpha (eels and tarpons) and Osteoglossomorpha (bony tongues) are each others closest relatives.
2022
November 2022
Fish Facility Manager Gia Haddock joins the MSU Fisheries & Wildlife Master's Program and Kristen Lounsbury joins us to take over in the fish room.
October 2022
|
The 10th Aquatic Models of Human Disease (AQMHD) Conference, co-organized by Ingo, Frauke Seemann (Texas A&M-Corpus Christi), Matt Harris (Harvard Medical School), and Patricia Schneider (Louisiana State University) was a great success!

|
Thanks everyone for coming and see you at the next AQMHD in San Antonio 2024!
August 2022
Two special members of our lab family move on to new positions:
|
Andrew Thompson opens his new Xtremo-Devo Lab as Assistant Professor at Western Michigan University. Congrats to Professor Drew at WMU in Kalamazoo!
Rachel Alcorn starts her Master's in Marine, Earth and Atmospheric Sciences at North Carolina State University. Good luck and thanks for all the fish! |
July 2022
|
Olivia, Drew, and Ingo host the Satellite Symposium Genomic Basis of Developmental Evolution in Chordates at the Joint Society for Developmental Biology - Pan-American Society for Evolutionary Developmental Biology Meeting in Vancouver.
Check out our exciting line-up out speakers and model systems and come to the SDB-PASEDB Meeting! |
February 2022
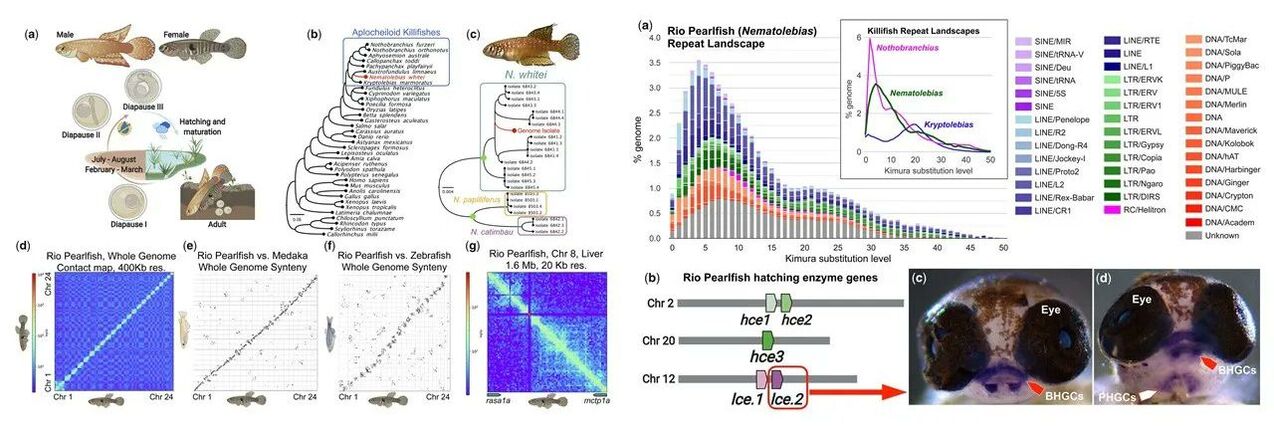
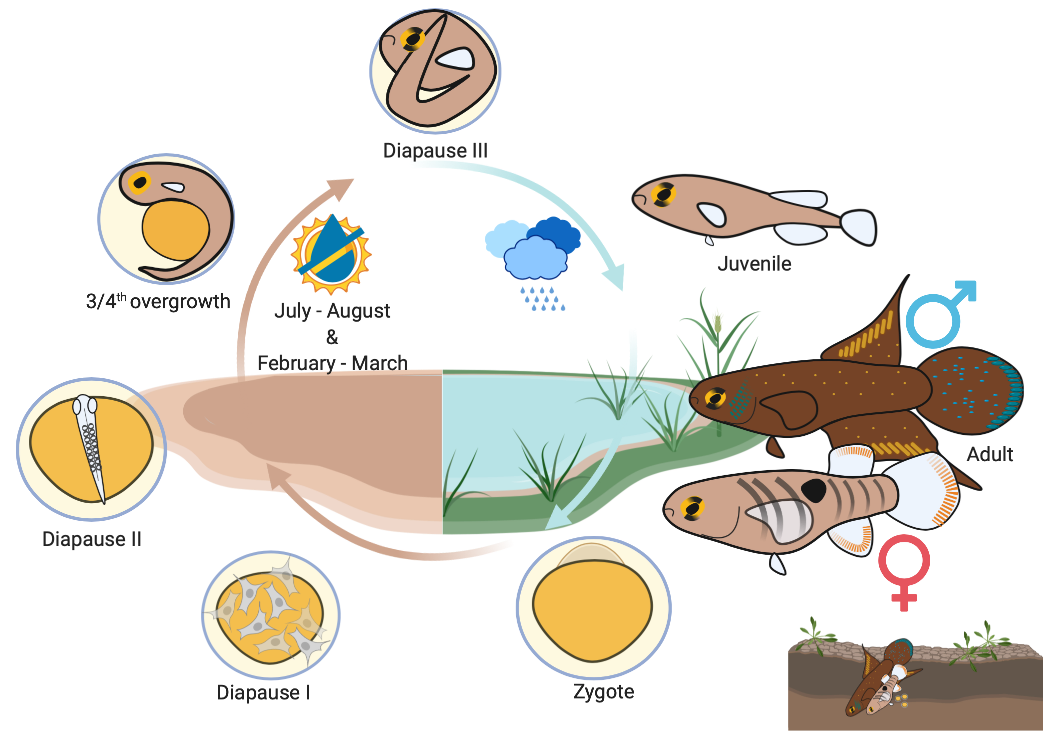
The Rio Pearlfish genome article is published in G3 Genes|Genomes|Genetics! Read the MSU Today story here.
Thompson et al. (2022) Genome of the Rio Pearlfish (Nematolebias whitei), a bi-annual killifish model for Eco-Evo-Devo in extreme environments
February 2022
See Ingo's talk on the Bowfin Genome project from the Genomes of Animals & Plants Virtual Conference.
January 2022
We have a new Zebrafish Facility Manager: Gia! Thanks to Taylor for all the good work keeping our facility afloat during the pandemic!
Three new undergraduate researchers are joining the team: Daniel, Chloe, and Keyana.
2021
October 2021
Our lab manager retires. Thanks for everything, Camilla - we will miss you!
August 2021
|
Our bowfin genome article is out!
Thompson et al. (2021) The bowfin genome of the bowfin (Amia calva) illuminates the developmental evolution of ray-finned fishes. Nature Genetics Read more about the story in MSU Today. See Bowfin also in the OneZoom Tree of Life Explorer. |
August 2021
Three new PhD students join the lab - our international lab is growing!
Welcome Jamily, Brooke, and Hao!
Welcome Jamily, Brooke, and Hao!
August 2021
Check out Cameron's interview with The Sci-Files on zebrafish neural crest Evo-Devo:
July 2021
Thanks to NSF IOS for supplemental funding to support post-baccalaureate student Rachel Alcorn over the next year to work on garnomics!
June 2021
Cameron leaves for graduate school at University of Colorado Anschutz. So long, and thanks for all the fish!
May 2021
Undergraduate Research Assistants Cameron and Rachel graduate - congrats to your achievement awards! We're glad you stay on to push our science forward!
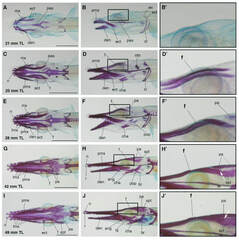
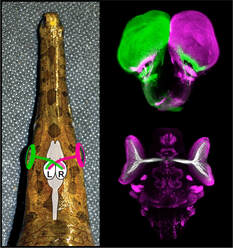
April 2021Gar SkullIn a new study in collaboration with the group of Tetsuya Nakamura published in Developmental Dynamics, we explore the development of the gar skull.
Gar Eyes
In a new study led by colleagues Alain Chédotal and Filippo Del Bene published in Science, we show that gar and other non-teleost fish have bilateral visual connections from eye to brain, revising the evolutionary history of 3D vision.
Read about it here in MSU Today. |
2020
December 2020
|
Our chromosome-level genome assembly plus NCBI RefSeq gene annotations for the Rio Pearlfish (Nematolebias whitei) are now publicly available.
See also our Killi-Kits, a STEM outreach tool that uses Pearlfish and other killifishes. |
November 2020
We received a $1.6 Mio. NSF IOS EDGE grant together with our collaborators Allyse Ferrara and Solomon David from Nicholls State University! More info here via MSU Today and NSF IOS.
October 2020
- The lab submitted five abstracts for #SICB2021 - see you online in January!
- Our new website is online. Hello World!